In contrast to the simplified molecular method the full molecular method treats a molecular ion rigorously. Impurity distributions for all atom species contained in the molecule are calculated, which enables to study pollution effects resulting from atom species that are implanted besides the doping species.
A fundamental assumption of the full molecular method is that the molecule dissipates immediately when it enters the target. This is true if the ion energy is much higher than the dissociation energy of the molecule which is of the order of 10 eV/atom. For ion energies in the keV-range this prerequisite for spontaneous dissociation is certainly met as long as the size of the molecule is not too big.
If this requirement is met the trajectories of all atoms of the molecule can be calculated separately without considering the motion of the other particles of the molecule, because the probability for a collision of two moving particles is low enough for neglecting such collisions. The various atoms of the molecule only interact with each other and with atoms of previously implanted molecules by the damage that is produced when the atoms move through the target.
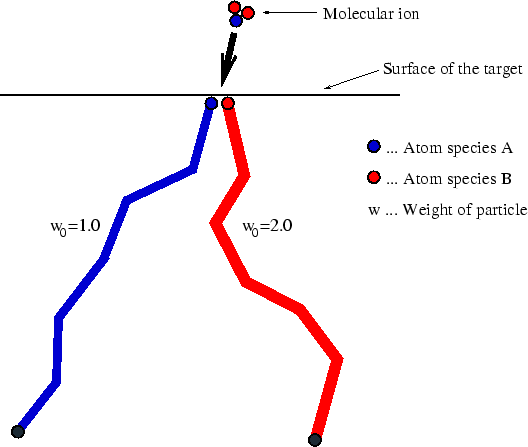
If the molecule contains more than one atom of the same species, these atoms
behave statistically identical. Therefore just one trajectory for each atom
species has to be calculated, whereas all other atoms of the same species are
taken into consideration by a weighting factor as indicated in
Fig. 4.9 for a molecule like BF![]() . The damage generation rate
and the concentration of the dopant species are multiplied with the number of
atoms of the same species in the molecule.
. The damage generation rate
and the concentration of the dopant species are multiplied with the number of
atoms of the same species in the molecule.
 |
Concerning damage recombination the full molecular method can be combined as well with the empirical damage model as with the Follow-Each-Recoil method. If the simulation is performed with the modified Kinchin-Pease model the same recombination parameters as if each atom species were implanted individually are applied.
![]()
![]()
![]()
![]() Previous: 4.5.2.1 Simplified Molecular Method
Up: 4.5.2 Molecular Ion Implantation
Next: 4.5.3 Point Response Interface
Previous: 4.5.2.1 Simplified Molecular Method
Up: 4.5.2 Molecular Ion Implantation
Next: 4.5.3 Point Response Interface