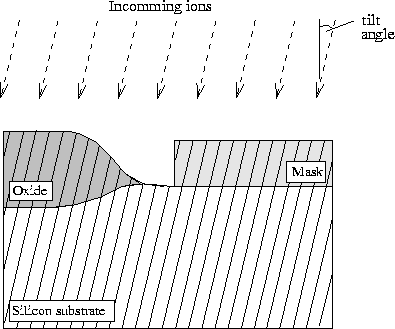
Figure 2.2-2: Discretization of the simulation geometry for a given tilt angle using the slab method. For the initialization of the vertical distribution function the numerical range scaling is applied.

Figure 2.2-2: Discretization of the simulation geometry for a given tilt angle
using the slab method. For the initialization of the vertical distribution
function the numerical range scaling is applied.
During the initialization of the vertical density function the flight path of the ions is assumed straight forward following the incoming direction. All intersections with the geometry are detected and used for the calculation of the characteristic parameters of (2.2-1) to (2.2-3). The algorithm can even handle holes or vacuum regions. In this case the initialization is continued when the ions hit the next non-vacuum region. We suppose implicitly that the ions do not change their direction when leaving the upper part of one layer and entering the lower part of the underlying one.
The same numerical algorithms are applied for the initialization of the lateral density function. Though lateral dopant profiles are hard to measure, their importance increases with the continuous decrease in minimal device dimensions. Moreover, it is of vital importance for the subsequent annealing process to obtain accurate initial conditions after the ion implantation. The simple Gaussian distribution is often used to model the lateral spread of the profiles [Gil88] [Fur72]. Monte Carlo simulations indicate that modified Gaussian density functions are well suited to model the lateral spread [Hob87] [Hob88].
To get the final concentration C(x,y) at a location (x,y), we add up the lateral and vertical distribution density functions by (2.2-5).
The convolution is performed in the normal coordinate system of the slices,
and the result is mapped onto the simulation grid point (x,y) by a
coordinate transformation. Normally, the simulation domains are always a
representative section of the real physical wafer domain, unfortunately, we
have to introduce artificial simulation boundaries. In reality we would have
a constant implantation profile at this artificial boundaries for a planar
wafer. Due to our convolution method the dopant concentration would drop off
at these boundaries, because the convolution integral is performed till
these artificial boundary and ,hence, the infinite tail is of the
convolution (2.2-5) is neglected. The loss of dopants will be
![]() at the boundary. By using a Gaussian lateral density function,
this artificial boundaries can be handled by mathematical expansions of the
integral to infinity (erfc-functions). This is only valid for non-tilted
(ion-beam perpendicular to surface) implantations. The only way to fulfill
the boundary conditions for tilted implants is to extend the simulation
geometry several times
at the boundary. By using a Gaussian lateral density function,
this artificial boundaries can be handled by mathematical expansions of the
integral to infinity (erfc-functions). This is only valid for non-tilted
(ion-beam perpendicular to surface) implantations. The only way to fulfill
the boundary conditions for tilted implants is to extend the simulation
geometry several times ![]() , where
, where ![]() is the standard
deviation of the target border material.
is the standard
deviation of the target border material.
As the resulting dopant concentration has to be defined on a simulation grid for further simulation tools, it is obvious that the density of the initial slices is determing the accuracy of the result. By increasing the slice density the simulation geometry is traced more efficiently. It should be pointed out at this place, that the simulation grid is only used as vehicle to transfer the information for subsequent simulation tools. For sake of simplicity we used an ortho-product grid, but there is no principal limitation for using unstructured grids.